Bumblebee
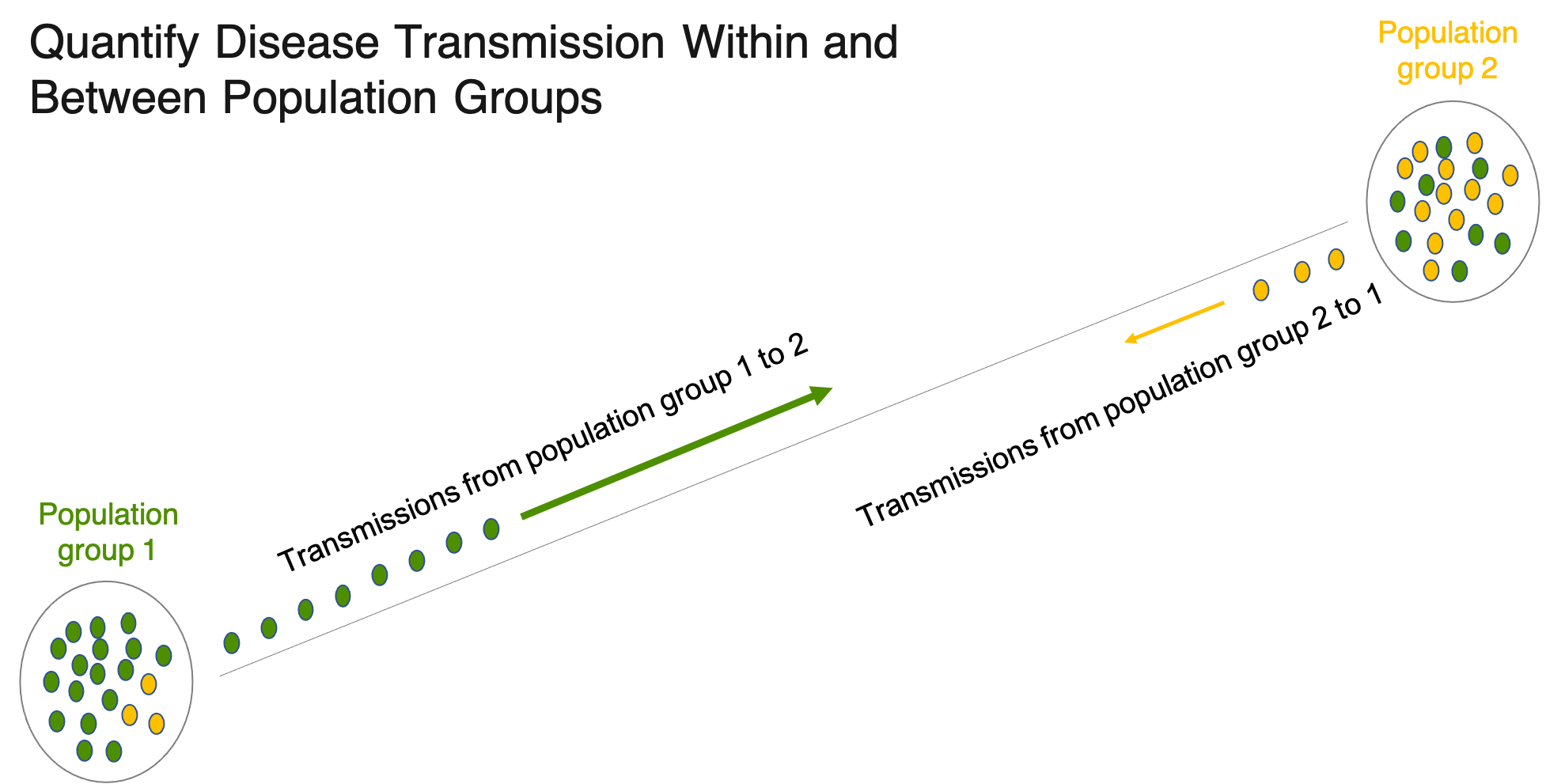
Overview
Bumblebee uses counts of directed transmission pairs identified between samples from population groups of interest to estimate the flow of transmissions within and between those population groups accounting for sampling heterogeneity.
Population groups might include: communities, geographical regions, age-gender groupings or arms of a cluster-randomized trial.
Why is this useful?
Quantifying the contribution of interventions, demographics and geographical factors to shaping patterns of transmission is important for understanding and ultimately controlling the spread of infectious disease.
Specifically, to address the following questions:
-
Which sub-populations are most at risk for infection and by whom?
-
Where are individuals most likely to acquire infection?
-
Are interventions having an impact on reducing disease spread?
Example application areas include:
-
Quantifying transmission patterns of HIV, the virus that causes AIDS, in the context of HIV prevention initiatives such as universal test-and-treat.
To learn more see: Magosi LE, et al., Deep-sequence phylogenetics to quantify patterns of HIV transmission in the context of a universal testing and treatment trial – BCPP/ Ya Tsie trial. eLife, 2022. 11:e72657. DOI: https://doi.org/10.7554/eLife.72657
-
Quantifying transmission patterns of SARS-COV-2, the virus that causes COVID-19, in the presence of heterogeneous vaccine uptake.
# To install the release version from CRAN:
install.packages("bumblebee")
# Load libraries
library(bumblebee) # for estimating transmission flows and confidence intervals
library(dplyr) # for working with data.frames
# To install the development version from GitHub:
# install devtools
install.packages("devtools")
# install bumblebee
library(devtools)
devtools::install_github("magosil86/bumblebee")
# Load libraries
library(bumblebee) # for estimating transmission flows and confidence intervals
library(dplyr) # for working with data.frames
Usage
Load the bumblebee package in your current R session, and try some examples in the example workflow
# Load libraries
library(bumblebee) # for estimating transmission flows and confidence intervals
library(dplyr) # for working with data.frames
# For an overview of available functions in bumblebee, type at the R prompt:
?estimate_transmission_flows_and_ci()
Details
To estimate transmission flows, that is, the relative probability of transmission within and between population groups accounting for variable sampling among those population groups, the bumblebee package computes the the conditional probability that a pair of individuals is from a specific population group pairing given that the pair is linked.
To read up about the statistical theory for estimating transmission flows see:
Magosi LE, et al., Deep-sequence phylogenetics to quantify patterns of HIV transmission in the context of a universal testing and treatment trial – BCPP/ Ya Tsie trial. eLife, 2022. 11:e72657. DOI: https://doi.org/10.7554/eLife.72657
Getting help.
To suggest a new feature, report a bug or ask for help, please provide a reproducible example at: https://github.com/magosil86/bumblebee/issues. Also see reprex to learn more about generating reproducible examples.
Code of conduct.
Contributions are welcome. Please observe the Contributor Code of Conduct when participating in this project.
Citation.
Magosi LE, et al., Deep-sequence phylogenetics to quantify patterns of HIV transmission in the context of a universal testing and treatment trial – BCPP/ Ya Tsie trial. eLife, 2022. 11:e72657 DOI: https://doi.org/10.7554/eLife.72657
References.
-
Magosi LE, et al., Deep-sequence phylogenetics to quantify patterns of HIV transmission in the context of a universal testing and treatment trial – BCPP/ Ya Tsie trial. eLife, 2022. 11:e72657. DOI: https://doi.org/10.7554/eLife.72657
-
Carnegie, N.B., et al., Linkage of viral sequences among HIV-infected village residents in Botswana: estimation of linkage rates in the presence of missing data. PLoS Computational Biology, 2014. 10(1): p. e1003430.
-
Goodman, L. A. On Simultaneous Confidence Intervals for Multinomial Proportions Technometrics, 1965. 7, 247-254.
-
Cherry, S., A Comparison of Confidence Interval Methods for Habitat Use-Availability Studies. The Journal of Wildlife Management, 1996. 60(3): p. 653-658.
Authors.
Lerato E. Magosi and Marc Lipsitch.
Maintainer.
Lerato E. Magosi lmagosi@hsph.harvard.edu or magosil86@gmail.com
License.
See the LICENSE file.

